Barcoding Sussex
DNA 'Barcoding' is the process of extracting and amplifying a short region of DNA from a specimen in order to apply it with a unique molecular 'barcode' that can be used to identify collections and determine species boundaries.
Fresh or preserved fungal tissue is pulverised and immersed in a lysis solution to extract its DNA, which is then transferred to a tube containing Taq polymerase. This is then cycled in a thermocycler to amplify the targeted DNA region into millions of copies. This process is called 'PCR'-testing (Polymerase Chain Reaction).
To check whether the process has successfully amplified the targeted DNA region, agarose gel electrophoresis is employed, which is a method of separating out and visualising the PCR products through an electrically-charged block of gel. The products are then compared to a reference known as a DNA 'ladder' to make sure the products we have generated are of the right length and are free of contamination.
PCR products are then sent via post to a sequencing facility in exchange for DNA sequences of our collections. The process from collection, through extraction and amplification to sequencing can be as little as five days.
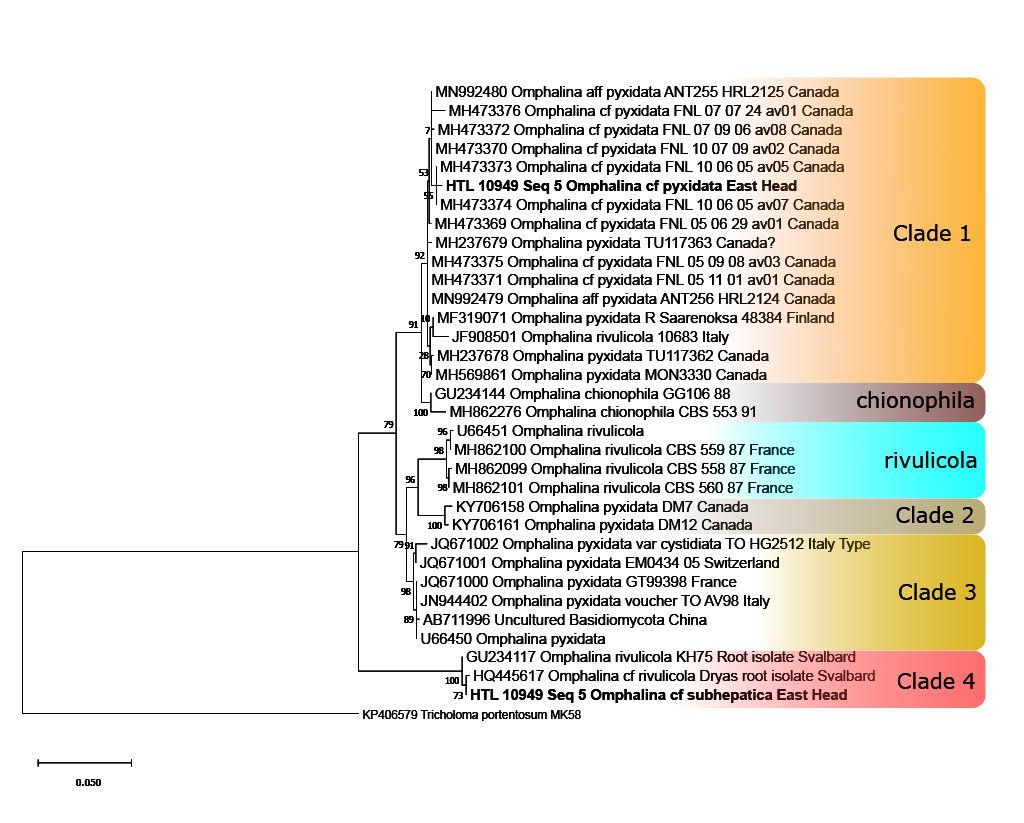
With the help and support from scientists at Royal Botanic Gardens, Kew, and partial funding by The British Mycological Society, we have been able to autonomously and successfully sequence many dozens of Sussex fungal specimens, confirming new species to our region and even Country.
Our molecular work will underpin our other projects and will be continuously submitted to online molecular databases such as Genbank in order to contribute to our knowledge of the Fungal Kingdom.
We have learnt how to edit, analyse and align DNA sequences in order to compare them. These alignments have enabled us to create robust phylogenetic trees, visualising the relationship between our collections and others submitted to online public databases.